?
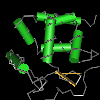 
ARID/BRIGHT DNA binding domain Members of the recently discovered ARID (AT-rich interaction domain) family of DNA-binding proteins are found in fungi and invertebrate and vertebrate metazoans. ARID-encoding genes are involved in a variety of biological processes including embryonic development, cell lineage gene regulation and cell cycle control. Although the specific roles of this domain and of ARID-containing proteins in transcriptional regulation are yet to be elucidated, they include both positive and negative transcriptional regulation and a likely involvement in the modification of chromatin structure. The basic structure of the ARID domain domain appears to be a series of six alpha-helices separated by beta-strands, loops, or turns, but the structured region may extend to an additional helix at either or both ends of the basic six. Based on primary sequence homology, they can be partitioned into three structural classes: Minimal ARID proteins that consist of a core domain formed by six alpha helices; ARID proteins that supplement the core domain with an N-terminal alpha-helix; and Extended-ARID proteins, which contain the core domain and additional alpha-helices at their N- and C-termini. |
