NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
NCBI Introduces PubReader, a New View for Full-Text Articles
Wednesday, December 19, 2012
As announced recently in the National Library of Medicine’s Technical Bulletin, NCBI now offers PubReader – a new, reader-friendly display format for full-text articles in the PubMed Central (PMC) literature database.
Leveraging the features of the latest underlying web technologies (HTML5 and CSS3), PubReader addresses usability and readability challenges specific to viewing research articles on tablets and other small-screen devices. PubReader also works on desktop and laptop computers.
Any article that is available in full-text HTML in PMC is viewable in the PubReader format. Furthermore, the PubReader functions with any of the latest browsers without the need to download an app or any additional software.
Like a printed journal article, PubReader breaks a document into multiple columns and pages to improve readability and navigation. PubReader can expand a page to whatever fits your screen, with multiple columns on a desktop monitor or a single column page on a small tablet. It will even switch to two columns if you rotate the tablet to a landscape view. When you adjust the font size or change the size of the browser window, page boundaries and columns adjust automatically.
The PubReader presentation shown in Figure 1 offers a variety of common options for moving between pages. You can use the PageUp, PageDown, RightArrow, LeftArrow keys on a keyboard, tap or click in the right or left margin (Figure 1A), use finger swipes on a touch screen device, or use the progress bar (Figure 1B) at the bottom of the screen to jump across the page range.
There is an image strip (Figure 1C) at the bottom of the PubReader page with thumbnails of all figures and tables in the article, allowing you to pop up an earlier figure/table and then close it in an instant without losing your place in the article. This same feature works with inline figures, as well as tables and citations. You will discover that PubReader has a number of other features to improve your reading experience as you use it in PMC.
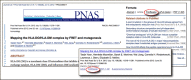
You can read more about the PubReader view on the PubReader about page. You can try it directly with an example record (PMCID: 3396517) or by clicking on the “PubReader” link for an article in a PMC search result list or in the article itself as shown in Figure 2.
Figure
Figure 2. Accessing the PubReader from standard record views in PMC. The link is in the Formats section in the full PMC article (Top Panel) and under the summary in search results (Bottom Panel).
The CSS and JavaScript code used to create the PubReader display are freely available from NCBITools on the public code repository GitHub. Anyone can use or adapt this code to display journal articles or other content that is structured as an HTML5 document.
GenBank Release 193.0 is Available
Monday, December 17, 2012
The new release for GenBank is now available via ftp.ncbi.nlm.nih.gov, as well as in the Nucleotide database and Blast services.
In release 193.0 (12/12/2012), the total number of non-WGS, non-CON records was 161,140,325 comprised of 148,390,863,904 basepairs of sequence data. In addition, there were 92,767,765 WGS records containing 356,002,922,838 basepairs of sequence data.
During the 65 days between the close dates for GenBank Releases 192.0 and 193.0, 830,113 records were updated with an average of 62,780 non-WGS/non-CON records added and/or updated per day.
- The non-WGS/non-CON portion of GenBank grew by almost 3 billion basepairs and by more than 3.25 million sequence records
- The WGS component of GenBank grew by over 22 billion basepairs and by more than 6.3 million sequence records
The total number of sequence data files increased by 45 with this release, with the divisions that expanded in file number:
- BCT = 3 new files, now a total of 94
- CON = 3 new files, now a total of 179
- EST = 3 new files, now a total of 469
- GSS = 6 new files, now a total of 166
- PAT = 4 new files, now a total of 186
- PLN = 1 new file, now a total of 60
- ROD = 1 new file, now a total of 30
- TSA = 21 new files, now a total of 133
- VRL = 2 new files, now a total of 24
- VRT = 1 new file, now a total of 28
The total number of AUT (author name) index files increased by 4 with this release and is now composed of 106 files.
For downloading purposes, please keep in mind that these GenBank flatfiles are roughly 579 GB (sequence files only) or 624 GB (including the 'short directory', 'index' and the *.txt files) when uncompressed. The ASN.1 formatted datafiles are approximately 474 GB.
For additional release information, see the Release Notes and README files in individual directories.
Now in PubChem: 8+ million Patented Chemicals from the SureChem Database
Monday, December 10, 2012
The SureChem patent chemistry database has deposited chemistry information for their complete collection of US, EP and WO full text patents from the 1976 to the present in PubChem.
More than 8 million structures are now available in the PubChem Substance database. Over 4 million of these structures are new to the PubChem Compound database which greatly expands public access to novel medicinal chemistry. For more information about each substance, users can click on the External ID link listed on the PubChem Substance records to go to the corresponding record SureChem's website.
SRA Surpasses a PetaBase of Sequence Data
Tuesday, December 04, 2012
The SRA database now contains more than a PetaBase (1x10^15 bases) of sequence data, including more than 390 TeraBases of open-access data from INSDC partners NCBI, EBI and DDBJ and more than 610 TeraBases of controlled-access human clinical sequence data.
- NCBI News, December 2012 - NCBI NewsNCBI News, December 2012 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...